addons, orange25, pypi
The easy way to install add-ons
BIOLAB
Nov 30, 2012
The possibility of extending functionality of Orange through add-ons has been present for a long time. In fact, we never provided the toolbox for crunching bioinformatical data as an integral part of Orange; it has always been an add-on. The exact mechanism of distribution of add-ons has changed significantly in the last year to simplify the process for add-on authors and to make it more standards-compliant. Among other things, this enables system administrators to install add-ons system-wide directly from PyPi using easy_install or pip. Unfortunately there were also negative side effects of this process, notably the temporary breakage of the add-on management dialog within the Orange Canvas.
We are happy to report that this is now being taken care of and you are encouraged to test the functionality.
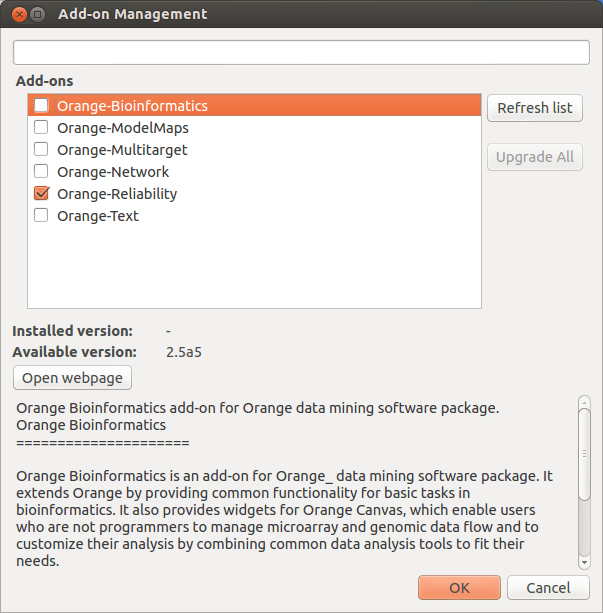
Select "Add-ons..." in the Options menu. A dialog will open that will list and describe existing add-ons. You can see the same list on the appropriate part of Orange website, but there is more. In the dialog, you can simply pick the add-ons you wish to use, confirm the selection and you should be good to go: widgets that come with the selected add-ons will become available immediately.
In case you change your mind, on some systems you can also uninstall add-ons by removing the check marks in front of them. This only works if you have pip installed, which is uncommon on Windows systems.
This might be a good time to warn you that the described functionality is new and not thoroughly tested on all the platforms on which Orange runs. If you stumble upon any strange or unwanted behavior, please let us now on the Orange forum, preferrably in the Bugs section.
Note that the Orange-Text add-on requires a compiler and appropriate libraries on your computer, and it as of now still refuses to be installed using the dialog. This is a known bug.